Apo-CGRP
Dataset
Full Case Study
Part One: Setup
Ensure you have created a location to keep track of CryoSPARC projects:
| Title | Value |
|---|---|
| Title | |
| Description | Leave empty |
| Directory | |
| Create Workspace | Enable |
| Workspace Title | Leave default |
There are a few prepared jobs that we will import into the project. They were exported from a different CryoSPARC instance and transferred onto the local SSD of your EC2 instance.
- Navigate to the project directory:
-
Make a new directory called
importsand navigate into it: - Move J188_nonuniform_refine_new
- Move J192_new_local_refine
- Move J719_volume_tools
Navigate to the workspace you just created, click on the dropdown next to 'New Job', select 'Import Job' and fill out the corresponding absolute path. Do this for all three jobs. You should see three completed jobs in the workspace:
| Import Path | Job Type | Imported Job Title |
|---|---|---|
| Non-Uniform Refinement | None | |
| Local Refinement | None | |
| Volume Tools | None |
- Input: particles from NU-refine job
- Input: mask from imported Volume Tools job
| Parameter | Value |
|---|---|
| Filter resolution (A) | |
| Cache particle images on SSD | False |
This job should take around 25 minutes to complete.
Create a 3D Variability Display job to visualize the results of the 3D Variability Analysis.
| Parameter | Value |
|---|---|
| Crop to size (after downsample) | |
| Filter resolution (A) | |
| Cache particle images on SSD | False |
This job should take around a minute to complete.
Download each component for both results and open them in UCSF ChimeraX.
Connect the volume from the imported Local Refinement job.
Use default parameters
The job should take ~4.5 minutes to complete.
Use the segger tool in ChimeraX to create a
.seg
file from the volume you just created. This will allow you to define the segments and connections for the 3D Flex job.
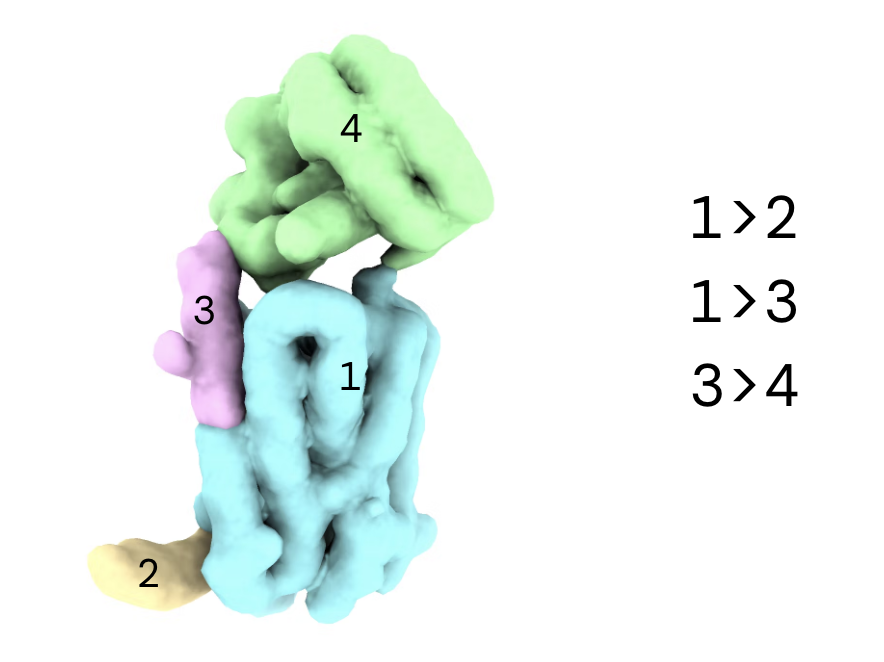
Drag and drop the
.seg
file into the browser. Ensure the correct project is selected and click 'Upload'. Once complete you can copy the absolute path of the file to use in the next step.
| Parameter | Value |
|---|---|
| Segmentation file path | |
| Segment connections | |
| Base num. tetra cells | |
| Min. rigidity weight | |
| Mask dilation (A) | |
| Mask threshold |
The job should take ~10 seconds to complete.
| Parameter | Value |
|---|---|
| Number of latent dims | |
| Rigidity (lambda) | |
| Latent centering strength | |
| Initialize latents from input | True |
Create a 3D Flex Generate job to visualize the results of the 3D Flex Training.
Input: results from 3D Flex Training job Input: volume from Non-Uniform Refinement job
Download the generated
.mrc
files and open them in UCSF ChimeraX.